New tool could help scientists understand brain structure changes underlying conditions such as autism

Scientists can now acquire detailed 3D microscopy images of an entire mouse brain in just hours thanks to technology advances such as the high-speed imaging technique known as light sheet microscopy. Although this new imaging data is providing incredible insights into the relationships between brain structure and disease, behavior and cognition, it also comes with some big analysis challenges.
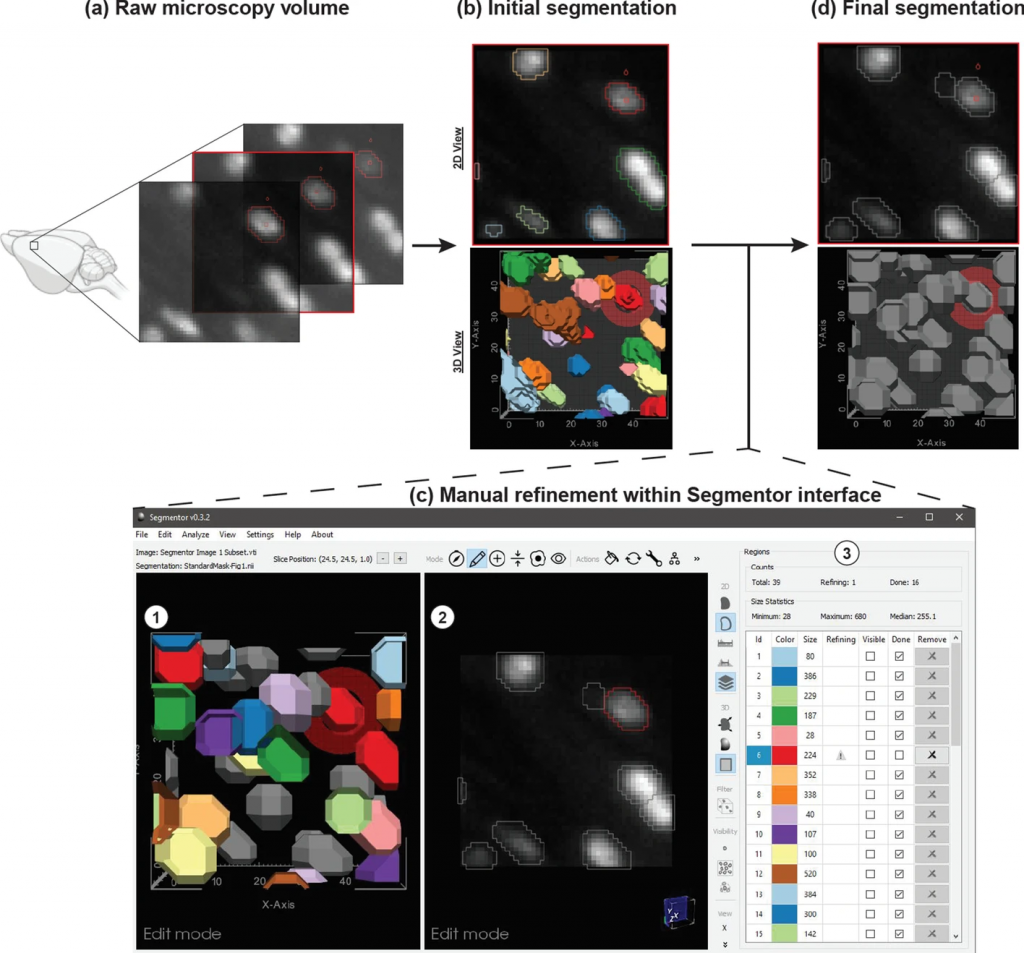
The images obtained with light sheet microscopy capture subcellular information for the approximately 100 million cells that make up the mouse brain. Making full use of this huge amount of data requires the daunting task of identifying important features such as nuclei in every cell. Although machine learning can help, algorithms must be trained to understand what a nucleus looks like, which requires large numbers of manually labeled nuclei to use as training data.
“Creating the training data is a challenging problem because the images can be noisy, and in some areas of the brain, the nuclei are packed so densely that it is hard to separate them out,” said David Borland, senior visualization researcher at RENCI and co-PI of the Nuclei Ninja project that is developing a high throughput platform for exploring and analyzing whole brain tissue cleared images. “To solve this problem, we developed the Segmentor software to produce high-quality data for training a machine learning algorithm to perform automatic segmentation.”
Acquiring training data
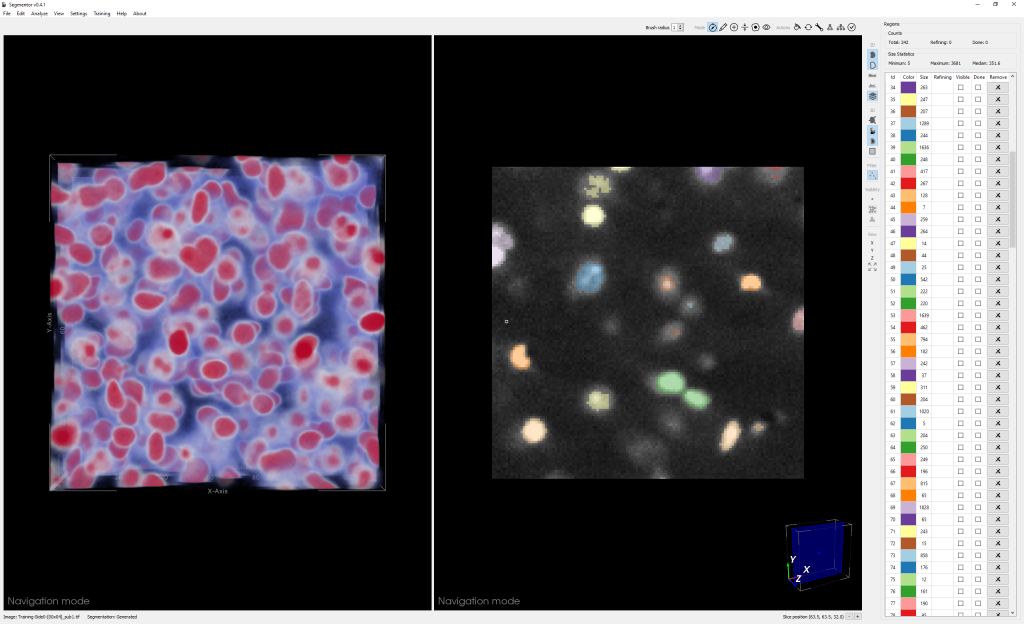
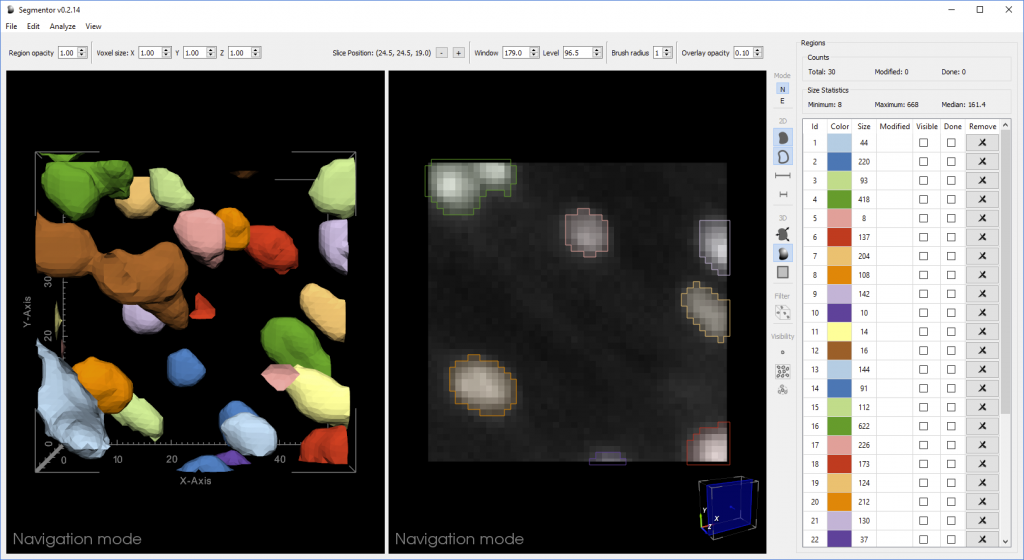
Segmentation of nuclei in brain images requires labeling all the 3D pixels, or voxels, that represent an individual nucleus in an image. The Segmentor software provides a very rough segmentation that a person can then refine. These refinements are fed back to the machine learning algorithm, which can use this input to produce segmentations that give users a better starting point for refining the segmentation next time. Once enough training data has been acquired, the algorithm should be able to produce segmentations that only need minimal corrections.
“This tool will eventually allow us to easily quantify differences in brain structure that are caused by genetic mutations associated with conditions that affect the brain, such as autism spectrum disorder,” said Borland. “And while we designed Segmentor for microscopy images, it can also be used with other imaging modalities such as MRI and CT.”
Light sheet microscopy produces a series of 2D image slices that are put together to create a large 3D image. One of the biggest challenges for creating Segmentor was figuring out ways for the user to intuitively interact with such complicated 3D data. The manual segmentation process must also be efficient because a single volume of data assigned for segmentation can contain thousands of nuclei.
Combining 2D and 3D visualization
“We created 3D visualization and 2D slice-based views that work in concert so that the user can edit in either view,” said Borland. “The 2D view reveals voxel intensities while the 3D view can make it easier to see the cellular geometry, which is especially useful if nuclei are close together.”
In a recent paper, the researchers showed that the Segmentor 2D-3D hybrid approach was two times faster than editing the same set of images with only 2D capabilities, without sacrificing accuracy. This increased efficiency could help the researchers more quickly reach their goal of acquiring around 20,000 high-quality manual 3D nuclei segmentations to train the machine learning algorithm.
A citizen science solution
To further ramp up the rate of acquiring training data, the researchers are working to turn their desktop tool into cloud-based citizen science software.
“We want to make an interface for people that is simpler and helps further distribute the work so that we can get more people contributing,” said Borland. “This is important because there are many areas of the brain, each with different characteristics. Even if we have enough training data for one part of the brain, we still need more training data for other parts of the brain to get good results for a whole brain.”
Other PIs for the Nuclei Ninja project include Guorong Wu and Jason L. Stein from UNC Chapel Hill and Minjeong Kim from UNC Greensboro.